While working on livestock management software for a client, I got to thinking. How much harder would inbreeding calculations be, if an organism could travel back in time to be its own ancestor? Disclaimer: this has not been peer reviewed, let alone reviewed by anybody with more than a high school level understanding of biology or time travel.
First, let’s start with an incredibly degenerate case. Alice takes her son Bob on a vacation into the future. While then, Bob picks a pretty flower and hides it in his pocket. He names the flower Carol. Upon returning to their native time, Bob immediately runs outside and replants the flower. Some time later, the flower fertilizes itself and produces an offspring, which is later picked up by young Bob.
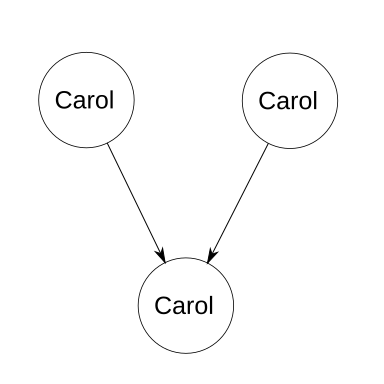
In this case, Carol contributed both of the haploids that merged to form herself. She’s her own parent, twice over. Each of Carol’s homozygous loci has a 100% chance of producing identical alleles in the gametes, and therefore the same homozygous locus in the offspring (which is also Carol). Each of Carol’s heterozygous loci has a 50% chance of giving the same allele to the male gamete as what was in the male gamete that produced Carol; similarly and independently, it has a 50% chance of giving the correct allele to the female gamete. Combined, there’s a 25% chance that a single heterozygous locus will result in the same genetic path that produced itself. However, unlike with normal inbreeding calculations, we know that all the genes of Carol as a child are equal to Carol as the parent. Therefore, let n be the number of loci, and the probability of heterozygosity due to descent is 0.25n. The probability of homozygosity due to descent, i.e., the coefficient of inbreeding, is therefore 1 − 0.25n. Luckily, Alice is a botanist, so when she discovers what Bob did, she teaches her son an interesting lesson about genetics and time travel.

Elsewhere in the world, Dave is a sheep farmer who’s about to go on a business trip to a few years in the past. Unfortunately, things don’t go according to plan, when his prized ewe Eve sees the portable time machine and thinks it’s a bucket of corn. She bumps into Dave, who was already nervous about time travel. He fumbles the time machine, which falls on Eve and activates. Dave spends the rest of the day talking to insurance companies. Meanwhile in the past, Eve meets a very attractive ram named Fred.
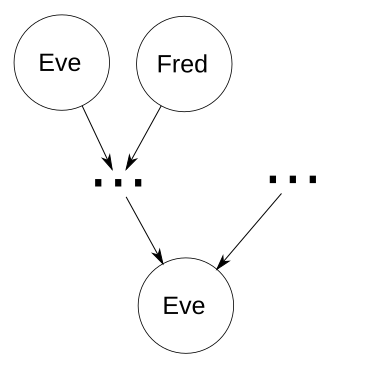
Since this is theory instead of reality, let’s ignore what actually happens next, and instead generalize and assume. Two of Eve’s ancestors are a mated couple, herself and Fred. Without looping, there are n generations from Eve as an ancestor to Eve as a descendant. I.e., n=1 represents Eve as her own parent, n=2 represents Eve as her own grandparent but not her own parent, etc. To make things easier, assume there are no other common ancestors, so that every ancestor of Eve that is not also a descendant of Eve has a coefficient of inbreeding of 0. Tracing genetic paths from Eve0 back n generations gives 2n distinct genetic paths, one of which is to Eve1 herself. Thus, each locus has a probability of 2−n+1 of having an allele passed down from Eve1 to Eve0. For each of those loci, if Eve1 is homozygous at the locus, she’ll pass down the correct allele (the one she got from herself, Eve2). If Eve1 is heterozygous at that locus, there’s a 50% chance she’ll pass down the correct allele. As in the degenerate case, we know that all of the genes that are passed down to herself are the same ones received from herself. So let m be the total number of loci, and the expected number of loci with alleles Eve passes to herself is 2−n+1m. Therefore, the probability of heterozygosity at those loci is 0.5(2−n+1m). The other loci have alleles from unrelated ancestors, so Eve’s total probability of homozygosity is 2−n+1(1 − 0.5(2−n+1m)). Except for small values of n, that’s really not bad at all!
Generalizing these cases into a comprehensive algorithm for calculating inbreeding in the face of time travel is left as an exercise for the reader. But I suspect it involves a multidigraph where each vertex represents an individual organism, and each arrow represents a gamete, pointing from the parent that created the gamete to the child created from the gamete. Then that type of graph can be split into cases, with some subgraphs analyzed separately. I think that would probably give an algorithm that’s a hybrid between the above cases and normal inbreeding calculations, but I’m not sure.












